Scientific Methodology and Work Packages
- Overall strategy and general description
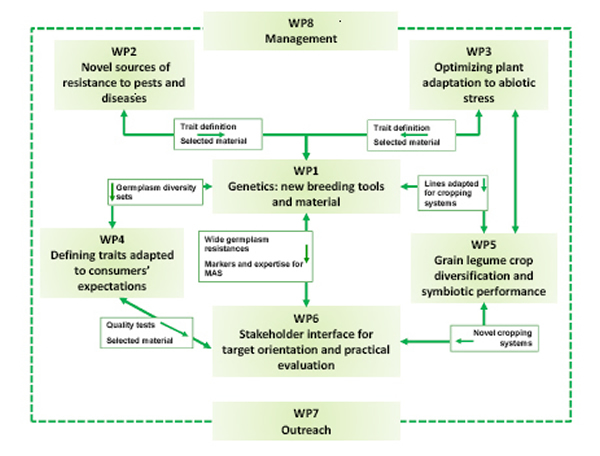
- Graphic diagram
- WP1: Genetics and new breeding tools and material (Lead: CRA)
- WP2: Biotic stress resistance (Lead: CSIC)
- WP3: Optimizing plant adaptation to abiotic stress (Lead: INRA)
- WP4: Defining traits adapted to consumers’ expectations (Lead: ITQB)
- WP5: Grain legume cropping system management (Lead: SLU)
- WP6: Stakeholder interface for targets orientation and practical evaluation (Lead: PGRO-RL)
- WP7: Outreach (Lead: IAMZ-CIHEAM)
- WP8: Management (Lead: IT)
Overall strategy and general description
As a baseline, we plan to develop tools and resources to enable the application of state of the art breeding methodology to the principal EU grain legume crops. The breeding tools will benefit from new data sources including our interaction with the Pea genome sequencing project (WP1). This will provide a much higher marker density than that hitherto available, and greatly accelerate plant breeding by speeding up introgression of favourable alleles of genes of interest. Due to the phylogenetic proximity of Pisum sativum, Vicia faba, and Lathyrus species, the data obtained for pea will be readily transferable to the other species under study. In parallel, and interacting synergistically with improved marker availability, wide genetic resources for these species will be tapped in the project (WP1). These will include: recombinant inbred line mapping populations, wide crosses between Pisum sativum and wild relatives (P. fulvum, P. elatius), white lupin (Lupinus albus) genetic resource collection, faba bean landaces, and wide crosses in grass pea (L. sativa x L. cicera).
By exploiting these genetic resources, we should get access to new sources of resistance to different pests and pathogens: (WP2), and robustness and symbiotic efficiency (WP3). The project will necessarily focus on a limited number of pathogens and pests (WP2), in order to make a significant impact, and to complement other ongoing studies. Our targets chosen represent major constraints in different climatic zones of Europe. In addition to major pests and pathogens (weevils, Ascochyta, Potyvirus) of worldwide distribution, we have chosen pathogens of regional importance, such as Aphanomyces in central Europe and Orobanche in the Mediterranean basin, for which available control methods are ineffective, and levels of resistance insufficient. Sitona weevils have also been relatively little studied in the past, necessitating the standardisation of pest damage protocols.
These studies will also be carried out under drought-stressed conditions, to select genetic material best able to cope with this stress (WP3). We will identify the loci controlling auto-fertility in faba bean, to guide the introduction of this character into more robust varieties that can set seed even in the absence of the insect pollinator (WP1). Whereas European pea and faba bean breeding has been mainly concentrated on developing varieties for livestock feed, LEGATO includes an exploratory work package devoted to traits needed to increase human consumption (WP4). We will evaluate consumer’s preferences, including their perception of the sustainability of legumes. We will identify accessions possessing favourable nutritional, organoleptic and/or processing quality traits for breeding programs, and devise rapid screening methods to accelerate the introduction of these traits. Although the development of new legume varieties is a priority, their successful exploitation will also depend on the adoption of new culture practices. We plan to use a novel modelling approach to devise and propose legume-based cropping systems adapted to each pedo-climatic zone, based on local consultation (MASC®). These cropping systems will then be tested in situ and evaluated for several properties (WP5).
Grain legumes fix atmospheric nitrogen in symbiosis with soil bacteria (Rhizobiaceae). The symbiotic interaction is bacterial strain-specific for a given host legume, and will not occur if the rhizobial strain concerned is not present in the soil. The rhizobial complement of the soil, and the effect of inoculation, will be studied for a network of different pedo-climatic zones within our project (WP5).
Promising pre-breeding material identified in WP1 through 5 will be transferred for testing in a network of pan-European trial sites to be further evaluated within cropping systems adapted to local conditions (WP6). Upcoming prebreeding material and new varieties, with the corresponding expertise, will be provided to and evaluated by, a network of plant breeders. The programme will also include tests with four plant breeders of marker-assisted selection on their material, using markers to emerge from the project.
LEGATO Work Breakdown Structure
WP1: Genetics and new breeding tools and material (Lead: CRA)
The effort in this work package will be divided between generic marker generation, in the framework of the pea genome sequencing programme, and the development of genetic material for analysis of developmental traits. This workpackage involves the identification, distribution and deployment of allelic variation. Allelic variation with respect to aspects of fertility will be examined - the duration of the flowering period in pea and the ability to self-pollinate without insect tripping of flowers in faba bean (autofertility). WP1 will develop genetic markers for traits studied in other WPs and specific antinutritional metabolites in two of our target species. The real gain achievable from marker assisted selection will be assessed. Of our target crop species pea, despite having the most advanced genetics, lacks some important resources which we will develop. We will relate genetic maps to sequence data and deploy novel variation from wild pea species in discrete segments using chromosome substitution lines. Finally we will characterize the allele distribution in the main species of lupin cultivated in Europe. The SME participants will provide locus specific assays (SFH) and multi locus assays (GXP). The Pea genome Sequencing project is an international consortium devoted to applying nextgeneration sequencing technology (NGS) to elucidate the large (ca. 4.5 Gb) P. sativum genomic sequence. LEGATO will provide highly dense bin mapping SNPs, using RAD markers (Baird et al.), obtained by re-sequencing a set of F2 individuals from a cross between the variety Caméor and a distantly related wild pea accession.
WP2: Biotic stress resistance (Lead: CSIC)
Crop germplasm screens (pea, faba bean, grass pea) will be used to identify novel sources of resistance against weevils (both Sitona and Bruchus) and aphids, and to Ascochyta blights using several locations. To complement existing fungal resistance genes, additional sources of resistance will be searched for, playing particular attention to pre-penetration stages, such as papilla-based resistance, and their association with gene candidates known to confer this type of resistance in other crops will be tested. The availability of NGS sequence data will be exploited to refine markers for resistances against Mycosphaerella pinodes, Aphanomyces euteiches and insect pests. Knowledge on potyvirus resistance available for pea will be extended and applied to other related Fabeae tribe legume crops. Where resistance is mapped as a QTL, candidate gene mapping within the QTL interval will be tested for their suitability as markers.
WP3: Optimizing plant adaptation to abiotic stress (Lead: INRA)
We will analyse the plant’s response to abiotic stress, primarily the interaction between drought and heat stresses. We will focus on the effect of abiotic stresses on the legume-Rhizobium symbiosis. Root properties will be studied using innovative non-destructive imaging techniques.The outcome of these studies will be twofold: prediction of ideotypes adapted to withstand drought, and identification of candidate genes and bacterial strains involved in the symbiotic plants’ adaptation to abiotic stress that can be used for selection.
WP4: Defining traits adapted to consumers’ expectations (Lead: ITQB)
The WP will develop methods for evaluating grain legume quality for human consumption and apply them to screen genetic resources of the major European culinary grain legumes. We will quantify a series of compounds affecting nutritional quality, health-beneficial properties, and organoleptic and processing qualities. The methods developed and data obtained will serve to improve screens for these traits in breeding programmes. This WP will also assess consumer preferences for a range of products based on grain legumes.
WP5: Grain legume cropping system management (Lead: SLU)
The first step will be an ex-ante assessment of new cropping systems to identify IC and VM potentially adapted to the local pedoclimatic zone. Once selected, the productivity, yield and stability of the cropping systems will be evaluated in field trials. In parallel, the effect of the CS on managing biotic and environmental stresses will be determined. In a subset of these trials, endogenous rhizobium populations will be quantified and analyzed. Highly efficient strains will be selected. The requirement for rhizobial inoculation by elite strains will be measured by comparing yields of inoculated and non-inoculated seed lots.
WP6: Stakeholder interface for targets orientation and practical evaluation (Lead: PGRO-RL)
The project will evaluate the potential impact of the new breeding material and cropping systems that are developed. One task in WP6 will embrace a stakeholder forum of plant breeders that identify priorities for trait selection and evaluate new data and material emerging from the project. A second major task will be trials by a Europe-wide network of plant breeders, both public and commercial, of genetic material and crop management regimes, and of marker-assisted selection protocols, all arising from the project.
WP7: Outreach (Lead: IAMZ-CIHEAM)
This WP will complement the stakeholder interface of WP6 in disseminating LEGATO results to a broad audience including end-users, consumers, policy makers, and selected educational instances such as agronomy schools and nutritionist bodies. In coordination with other interested bodies such as the Legume Society, and the inter-professional associations (UNIP, PGRO), the WP will utilize a wide range of communication media from a dedicated Website through brochures, newsletter, press releases, a training course and an international conference.
WP8: Management (Lead: IT)
The Management will steer the project so that it remains of high relevance irrespective of any changes that may occur in the area should they be scientific, technological, or environmental. At the operational level, WP8 is designed to ensure that the project will be progressing in conformity to the work plan in particular with regard to the progress, the milestones, the deliverables, as well as the planned resources. At the organisational level, WP8 aims to achieve maximum efficiency of the infrastructure setup to support the project with special attention paid to financial, logistics, information, and coordination issues and in terms of quality and conformity to EC rules and procedures.
